MLBBQ — Benchmarking Methods for Mapping Functional Connectivity in the Brain
MLBBQ (Machine Learning Benchmarking for Brain Connectivity Quantification) is a conceptual and methodological framework designed to evaluate, compare, and standardize methods for mapping functional connectivity (FC) in the brain. It combines neuroscience, machine learning, and data benchmarking principles to improve the reliability and interpretability of connectivity studies.
Overview
Functional connectivity refers to the statistical dependence between spatially separated brain regions, often inferred from signals such as functional magnetic resonance imaging (fMRI), electroencephalography (EEG), or intracranial recordings. Because brain connectivity analyses rely on diverse computational models, results can vary depending on methodology.
MLBBQ benchmarking provides:
- Standardized datasets and evaluation criteria.
- Comparisons of traditional statistical methods with modern machine learning approaches.
- Insights into model biases, scalability, and biological validity.
Benchmarking Functional Connectivity Methods in Brain Mapping
Study Overview and Methodology
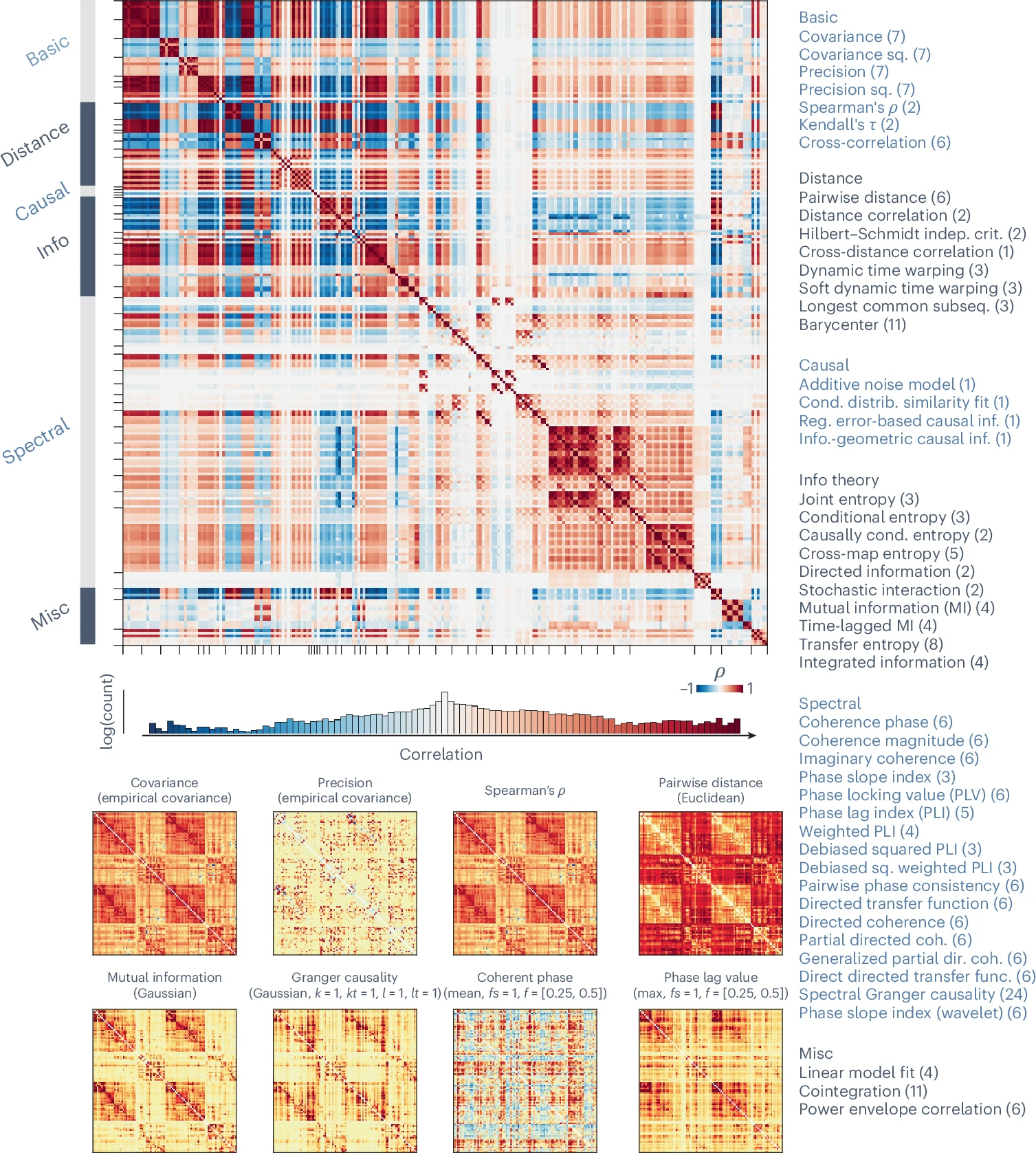
A large-scale benchmarking study assessed how different pairwise interaction statistics shape the organization and interpretation of functional connectivity networks.
- Dataset: Resting-state fMRI data from 326 healthy young adults in the Human Connectome Project (HCP).
- Methods: A library of 239 pairwise interaction statistics, derived from 49 measures across 6 methodological families, was applied using the Python Toolkit of Statistics for Pairwise Interactions (pyspi).
- Objective: Evaluate how the choice of pairwise statistic influences the topology of FC matrices and their relevance to neuroscience applications.
Impact on Network Organization and Biological Alignment
- Topological and Geometric Features
- Edge weights and hub ranking vary significantly across methods.
- Some methods localize hubs to unimodal cortex, while others distribute hubs along the unimodal–transmodal axis.
- Weight–Distance Relationship
- Most methods show an inverse relationship between FC magnitude and physical proximity.
- Strength of this relationship differs widely across statistics.
- Structure–Function Coupling
- Positive associations between structural connectivity (diffusion MRI) and FC are strongest for precision, stochastic interaction, and imaginary coherence.
- Alignment with Multimodal Biological Networks
- FC methods align differently with:
- Correlated gene expression.
- Laminar similarity.
- Neurotransmitter receptor similarity.
- Electrophysiological connectivity (MEG).
- Metabolic connectivity (FDG-PET).
- Strongest correspondences were found with receptor similarity and electrophysiological connectivity.
- FC methods align differently with:
Quantifying Individual Differences
- Individual Fingerprinting
- Precision-based statistics outperform Pearson correlation in identifying individuals from their FC patterns.
- Brain–Behavior Prediction
- Covariance, precision, and information-theory-based statistics best predict cognitive performance and lifestyle traits (e.g., tobacco use).
Information Flow Patterns
Using integrated information decomposition (ΦID), the study examined how FC methods capture different information dynamics:
- Redundant Information Storage
- Captured by classic methods (covariance, precision, mutual information).
- Diverse Information Dynamics
- Directed transfer function and partial coherence reveal information migration, duplication, encryption, and decryption.
Conclusions and Recommendations
- There is no single “optimal” FC method. The best choice depends on the research question and targeted neurophysiological mechanism.
- Covariance-based methods (especially precision/partial correlation) consistently perform well across criteria:
- Robust proximity–FC relationships.
- Strong coupling with structural connectivity.
- Alignment with biological similarity.
- High individual differentiation and predictive power.
This benchmark provides a comprehensive roadmap for researchers, emphasizing that statistical choices must align with biological hypotheses when studying brain connectivity.
Applications of MLBBQ Benchmarking
- Clinical Neuroscience: Biomarker discovery for neurological and psychiatric disorders.
- Cognitive Neuroscience: Mapping neural substrates of memory, attention, and perception.
- Neurotechnology: Optimizing brain–computer interfaces.
- AI and Neuroscience: Inspiring architectures for artificial neural networks based on connectivity principles.
Future Directions
- Development of standardized benchmark suites for FC (analogous to ImageNet/GLUE in AI).
- Integration of multimodal data (fMRI + EEG/MEG + genetics + PET).
- Wider adoption of explainable AI for neuroscientific interpretability.
- Expansion to dynamic connectivity mapping to capture temporal fluctuations.
Tags: MLBBQ, Benchmarking, Functional connectivity, Brain mapping, Machine learning, Neuroscience, Connectivity quantification, fMRI, EEG, Functional connectivity methods, Pairwise interaction statistics, HCP dataset, Human Connectome Project, Python Toolkit (pyspi), Precision correlation, Pearson correlation, Covariance methods, Brain network topology, Hub ranking, Weight–distance relationship, Structure–function coupling, Stochastic interaction, Imaginary coherence, Receptor similarity, Electrophysiological connectivity, MEG, Gene expression alignment, Laminar similarity, Metabolic connectivity, FDG-PET, Individual fingerprinting, Brain–behavior prediction, Information-theoretic statistics, Integrated information decomposition (ΦID), Directed transfer function, Partial coherence, Redundant information, Diverse information dynamics, Benchmark framework, Standardized datasets, Evaluation criteria, Model biases, Scalability, Biological validity, Clinical neuroscience, Cognitive neuroscience, Neurotechnology, Explainable AI, Dynamic connectivity, Multimodal data integration